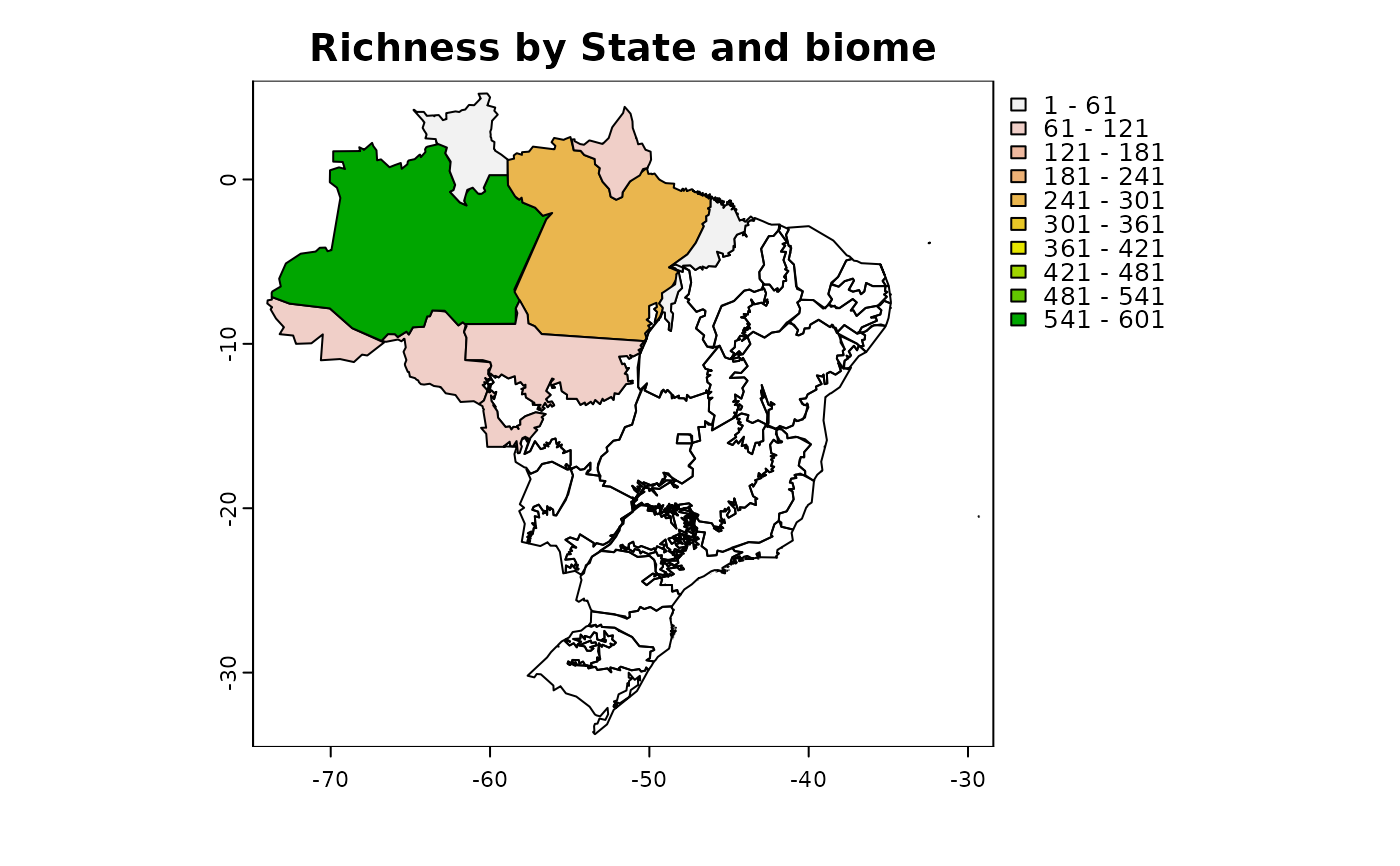
Get a presence-absence matrix of species based on its distribution (states, biomes and vegetation types) according to Flora e Funga do Brasil.
get_pam(data, by_biome = TRUE, by_state = TRUE,
by_vegetation = FALSE, remove_empty_sites = TRUE,
return_richness_summary = TRUE,
return_spatial_richness = TRUE,
return_plot = TRUE)Arguments
- data
(data.frame) a data.frame imported with the
load_florabrfunction or generated by eitherselect_speciesorsubset_speciesfunctions- by_biome
(logical) get occurrences by biome. Default = TRUE
- by_state
(logical) get occurrences by State. Default = TRUE
- by_vegetation
(logical) get occurrences by vegetation type. Default = FALSE
- remove_empty_sites
(logical) remove empty sites (sites without any species) from final presence-absence matrix. Default = TRUE
- return_richness_summary
(logical) return a data.frame with the number of species in each site. Default = TRUE
- return_spatial_richness
(logical) return a SpatVector with the number of species in each site. Default = TRUE
- return_plot
(logical) plot map with the number of species in each site. Only works if return_spatial_richness = TRUE. Default = TRUE
Value
If return_richness_summary and/or return_spatial_richness is set to TRUE, return a list with:
PAM: the presence-absence matrix (PAM)
Richness_summary: a data.frame with the number of species in each site
Spatial_richness: a SpatVector with the number of species in each site (only by State and biome)
If return_richness_summary and return_spatial_richness is set to FALSE, return a presence-absence matrix
References
Flora e Funga do Brasil. Jardim Botânico do Rio de Janeiro. Available at: http://floradobrasil.jbrj.gov.br/
Examples
data("bf_data") #Load Flora e Funga do Brasil data
#Select endemic and native species of trees with occurrence only in Amazon
am_trees <- select_species(data = bf_data,
include_subspecies = FALSE,
include_variety = FALSE,
kingdom = "Plantae",
group = "All", subgroup = "All",
family = "All", genus = "All",
lifeForm = "Tree", filter_lifeForm = "only",
habitat = "All", filter_habitat = "in",
biome = "Amazon",
filter_biome = "only",
state = "All", filter_state = "and",
vegetation = "All",
filter_vegetation = "in",
endemism = "Endemic", origin = "Native",
taxonomicStatus = "Accepted",
nomenclaturalStatus = "All")
#Get presence-absence matrix
pam_am <- get_pam(data = am_trees, by_biome = TRUE, by_state = TRUE,
by_vegetation = FALSE, remove_empty_sites = TRUE,
return_richness_summary = TRUE,
return_spatial_richness = TRUE,
return_plot = TRUE)